Genomic Profiling: Foundation for Personalized Therapy
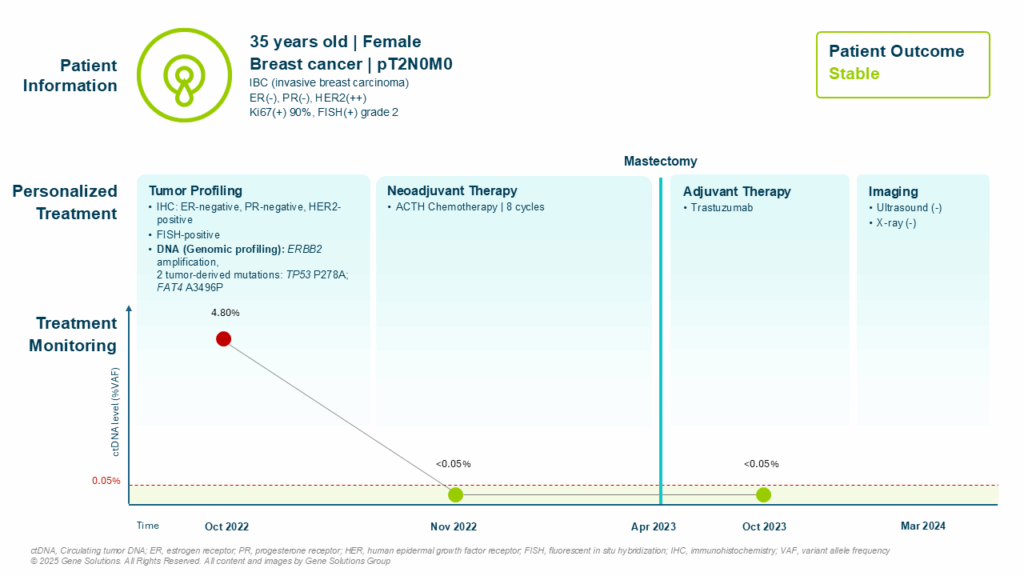
At initial diagnosis in September 2022, the patient presented with invasive breast carcinoma (ER-negative, PR-negative, HER2 2+ by IHC, confirmed FISH-positive). She had a Ki-67 index of 90% and was staged as pT2N0M0.
To gain deeper insight, her doctor ordered genomic profiling using the K-TRACK™ platform. The results revealed: ERBB2 amplification, confirming HER2 overexpression at the DNA level and two tumor-derived mutations: TP53 P278A and FAT4 A3496P, both associated with potential treatment resistance or aggressive tumor biology. (1,2,3)
These findings provided both diagnostic confirmation and prognostic insight, prompting the oncologists to initiate neoadjuvant HER2-targeted chemotherapy with close monitoring of treatment response.
ctDNA Monitoring: A Real-Time Biomarker for Tumor Dynamics
Prior to neoadjuvant chemotherapy, patient’s ctDNA analysis showed a variant allele frequency (VAF) of 4.80%, confirming the presence of circulating tumor-derived DNA. This molecular baseline allowed oncologists to dynamically track tumor response beyond imaging.
By the second cycle of therapy, ctDNA became undetectable, indicating a strong molecular response. Following completion of 8 cycles of systemic therapy, patient underwent mastectomy in April 2023. Histopathology revealed only residual ductal carcinoma in situ (DCIS) with no invasive component — consistent with a pathologic complete response (pCR).
Longitudinal ctDNA monitoring post-treatment showed sustained ctDNA negativity at both 12-month and 21-month follow-ups, supporting continued clinical remission and disease stability.
The Power of Combining Both Tools
This case underscores the combination role of genomic profiling and ctDNA monitoring:
- Genomic profiling enables comprehensive characterization of actionable mutations, prognostic markers, and molecular subtypes — guiding tailored therapy selection.
- ctDNA monitoring provides a dynamic and non-invasive means of evaluating treatment response, detecting minimal residual disease (MRD), and tracking long-term remission.
Reference:
- Huang, Y. et al. (2024) ‘TP53-specific mutations serve as a potential biomarker for homologous recombination deficiency in breast cancer: a clinical next-generation sequencing study,’ Precision Clinical Medicine, 7(2). https://doi.org/10.1093/pcmedi/pbae009.
- Hwang, S.H. et al. (2024) ‘Clinical Relevance of TP53 Mutation and Its Characteristics in Breast Cancer with Long-Term Follow-Up Date,’ Cancers, 16(23), p. 3899. https://doi.org/10.3390/cancers16233899.
- Liu, B. et al. (2022) ‘Molecular landscape of TP53 mutations in breast cancer and their utility for predicting the response to HER‐targeted therapy in HER2 amplification‐positive and HER2 mutation‐positive amplification‐negative patients,’ Cancer Medicine, 11(14), pp. 2767–2778. https://doi.org/10.1002/cam4.4652.