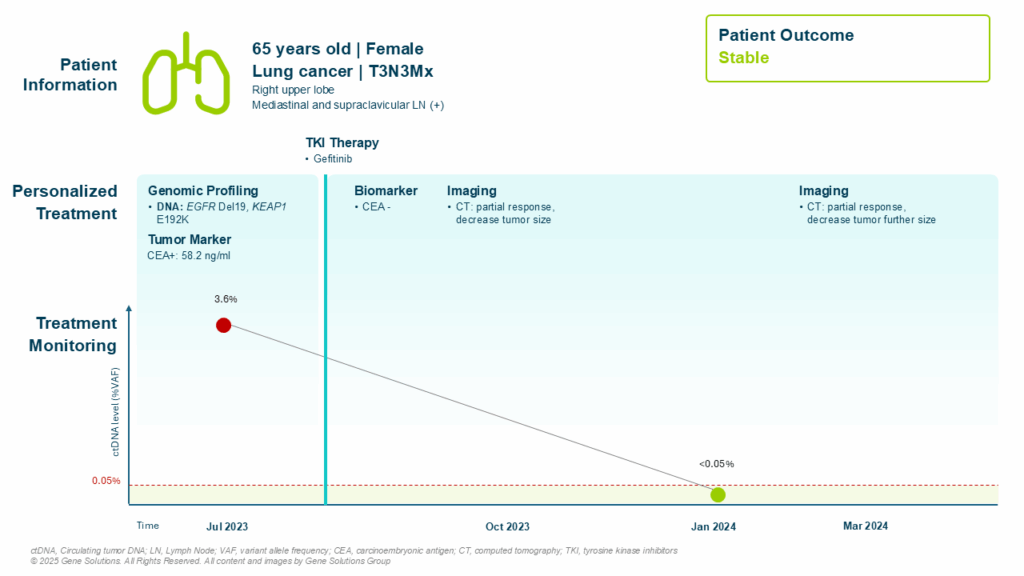
Genomic Profiling: Setting the Stage for Targeted Therapy
At diagnosis, Ms. L underwent tissue and plasma-based genomic profiling. The results revealed:
- EGFR exon 19 deletion, a well-characterized driver mutation predictive of response to first- and third-generation EGFR TKIs (1)
- KEAP1 E192K, a mutation increasingly associated with aggressive disease and potential resistance mechanisms in NSCLC (2)
These findings directly informed the decision to initiate gefitinib, a first-line EGFR inhibitor, offering a targeted alternative to cytotoxic chemotherapy. Importantly, genomic profiling also established a personalized molecular profile to monitor treatment efficacy through tumor-informed ctDNA monitoring.
ctDNA Monitoring: A Real-Time Indicator of Response
Baseline ctDNA analysis showed a variant allele frequency (VAF) of 3.60%, confirming the presence of circulating tumor DNA in plasma — a reflection of active tumor burden.
As treatment with gefitinib progressed, so did the molecular response:
- January 2024 (6 months post-treatment): ctDNA became negative (VAF < 0.05%), indicating molecular clearance
- This preceded continued imaging responses and reflected deep biological suppression of tumor activity
By March 2024, radiological imaging confirmed partial response and tumor size decrease, consistent with RECIST 1.1 criteria.
ctDNA was cleared after 6 months of treatment, corresponding with the partial response evaluated by RECIST 1.1 criteria at the 9-month visit, supporting its use as a dynamic biomarker for treatment response assessment.
This case demonstrates how these two tools operate on different yet synergistic levels:
- Genomic Profiling: Identifies therapeutic targets and resistance markers, Establishes a personalized molecular profiling for monitoring
- ctDNA Monitoring: Tracks response non-invasively and Offers real-time insight into tumor biology between scans
Reference:
- Truini A, Starrett JH, Stewart T, et al. The EGFR Exon 19 Mutant L747-A750>P Exhibits Distinct Sensitivity to Tyrosine Kinase Inhibitors in Lung Adenocarcinoma. Clinical Cancer Research. 2019;25(21):6382-6391. doi:10.1158/1078-0432.ccr-19-0780
- Hellyer JA, Stehr H, Das M, et al. Impact of KEAP1/NFE2L2/CUL3 mutations on duration of response to EGFR tyrosine kinase inhibitors in EGFR mutated non-small cell lung cancer. Lung Cancer. 2019;134:42-45. doi:10.1016/j.lungcan.2019.05.002