Initial Diagnosis: Actionable, Yet Complex
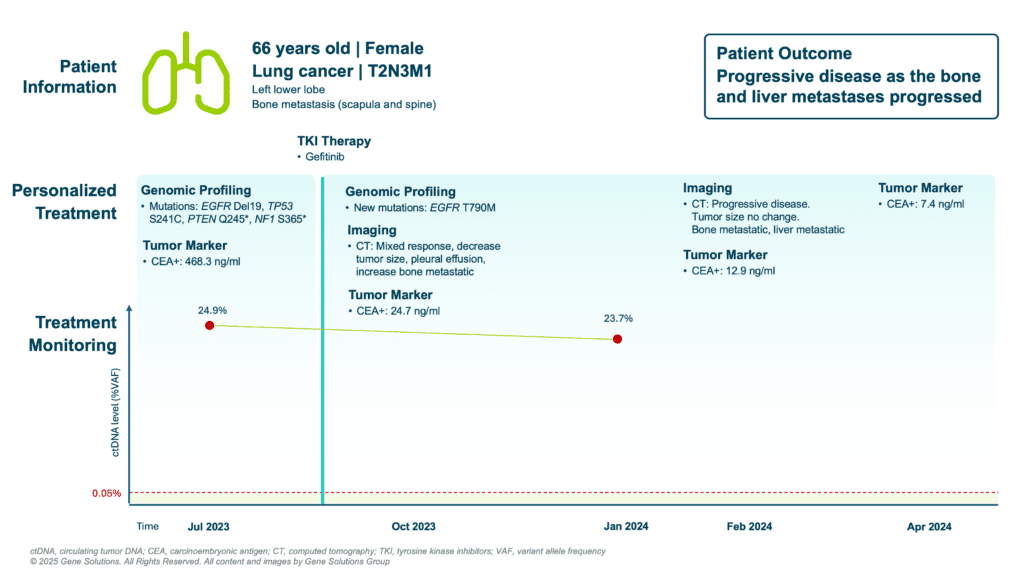
In July 2023, the patient was diagnosed with stage IV NSCLC (T2N3M1) involving bone metastases in the scapula and spine. Comprehensive genomic profiling of both tissue (FFPE) and plasma revealed multiple mutations:
- EGFR exon 19 deletion — a driver mutation predictive of response to EGFR TKIs (1)
- TP53 S241C, PTEN Q245*, NF1 S365* — co-mutations commonly associated with poor prognosis and heterogeneous treatment response (2,3)
These findings guided initiation of gefitinib, an EGFR tyrosine kinase inhibitor.
Genomic profiling enabled:
- Targeted therapy selection
- Early identification of potential co-mutation–related resistance risk
- Establishment of a personalized mutation profile for ctDNA monitoring
ctDNA Monitoring: The Story Beneath the Surface
At baseline, the patient’s ctDNA level was high (24.9%), and her CEA level was markedly elevated (468.3 ng/mL).
However, as treatment progressed:
- CEA dropped significantly — 24.7 ng/mL by October, and further down to 7.4 ng/mL by April 2024
- Yet, ctDNA remained persistently elevated — only slightly decreasing to 23.7% by January 2024
Critically, a new EGFR T790M mutation emerged in the ctDNA by October 2023, indicating acquired resistance to gefitinib — even before radiographic progression. While CEA appeared to reflect treatment success, ctDNA exposed persistent disease activity and molecular evolution.
Clinical Insight: What ctDNA Saw First
The case exemplifies how ctDNA can signal resistance earlier than imaging or serum markers:
- EGFR T790M, a common resistance mutation, was detected in ctDNA months before disease was deemed progressive by CT.
- Despite improving CEA, ctDNA captured true tumor biology, allowing oncologists to consider early intervention — potentially before clinical deterioration.
A Real-Time View Saves Real Time
This case reminds us that falling tumor markers aren’t always the full story. Persistent ctDNA and emergence of EGFR T790M reflected true tumor progression, enabling a window of opportunity for early treatment adjustment.
In an era where resistance is inevitable, ctDNA equips oncologists to respond before relapse becomes radiographically evident.
References:
- Gijtenbeek RGP, Damhuis RAM, Van Der Wekken AJ, Hendriks LEL, Groen HJM, Van Geffen WH. Overall survival in advanced epidermal growth factor receptor mutated non-small cell lung cancer using different tyrosine kinase inhibitors in The Netherlands: a retrospective, nationwide registry study. The Lancet Regional Health – Europe. 2023;27:100592. doi:10.1016/j.lanepe.2023.100592
- Canale M, Petracci E, Delmonte A, et al. Impact of TP53 Mutations on Outcome in EGFR-Mutated Patients Treated with First-Line Tyrosine Kinase Inhibitors. Clinical Cancer Research. 2016;23(9):2195-2202. doi:10.1158/1078-0432.ccr-16-0966
- Guo Y, Song J, Wang Y, et al. Concurrent genetic alterations and other biomarkers predict treatment efficacy of EGFR-TKIs in EGFR-Mutant Non-Small Cell lung cancer: a review. Frontiers in Oncology. 2020;10. doi:10.3389/fonc.2020.610923