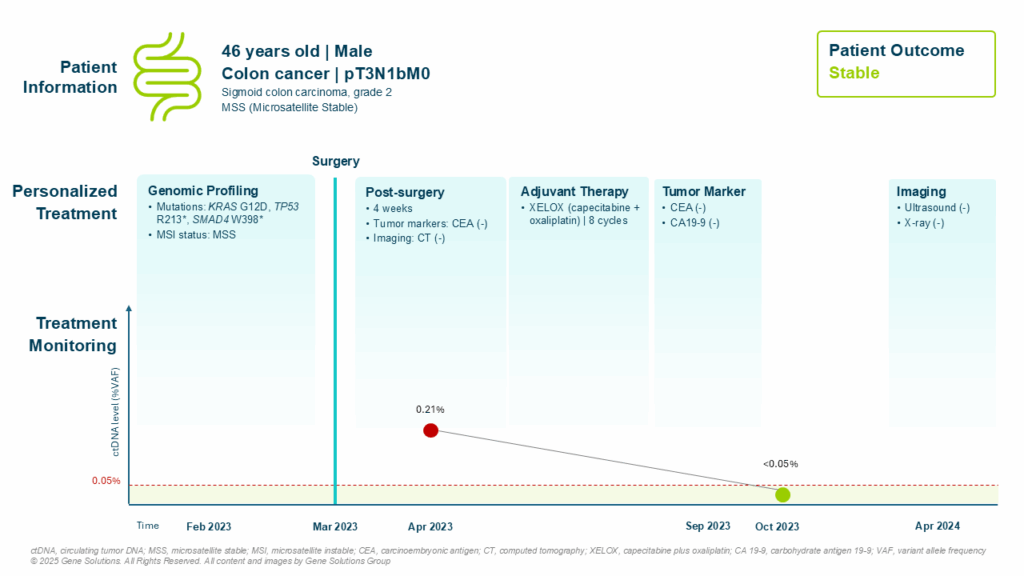
Genomic Profiling: Uncovering Hidden Risks
Post-surgical tumor profiling revealed three clinically relevant mutations:
- KRAS G12D – a well-known oncogenic driver, associated with poor response to anti-EGFR therapy (1)
- TP53 R213* – a loss-of-function mutation, often linked to genomic instability (2) and poor prognosis when combined with KRAS (3) or SMAD4 (4)
- SMAD4 W398* – commonly altered in advanced colorectal cancers, contributing to poor prognosis (5)
The combination of mutations can significantly alter prognosis. Research suggests that tumors harboring simultaneous KRAS, TP53, and SMAD4 mutations are associated with worse survival and recurrence (6), even when clinical staging suggests low risk. In his case, these findings prompted his oncologist to pursue a more proactive adjuvant therapy approach than might have been considered based on staging alone.
ctDNA Monitoring: Guiding Adjuvant Chemotherapy Decisions
At 4 weeks post-surgery, his CEA were negative — a reassuring sign using standard tumor markers. However, ctDNA testing revealed a positive result (0.21%), indicating the presence of minimal residual disease (MRD) not captured by imaging or serum biomarkers.
This ctDNA result directly influenced the decision to initiate adjuvant chemotherapy, and he began 8 cycles of XELOX (capecitabine + oxaliplatin) in April 2023.
ctDNA served as a real-time biomarker, identifying residual disease and justifying timely initiation of adjuvant chemotherapy — which may have prevented recurrence.
Positive Trajectory and Molecular Remission
Follow-up in October 2023 revealed ctDNA clearance, suggesting a favorable response to therapy. By April 2024, both imaging and tumor markers (CEA, CA19-9) remained negative, with no clinical evidence of recurrence.
In colorectal cancer — where adjuvant chemotherapy decisions can significantly impact outcomes — molecular tools like ctDNA and genomic profiling provide powerful, patient-specific guidance. In the case of this patient, this approach led to timely intervention and sustained remission.
Reference:
- Zhu, G. et al. (2021) ‘Role of oncogenic KRAS in the prognosis, diagnosis and treatment of colorectal cancer,’ Molecular Cancer, 20(1). https://doi.org/10.1186/s12943-021-01441-4.
- Li, W. et al. (2024) ‘Unraveling the role of TP53 in colorectal cancer therapy: From Wild-Type Regulation to Mutant,’ Frontiers in Bioscience-Landmark, 29(7). https://doi.org/10.31083/j.fbl2907272.
- Tang, Y. and Fan, Y. (2024) ‘Combined KRAS and TP53 mutation in patients with colorectal cancer enhance chemoresistance to promote postoperative recurrence and metastasis,’ BMC Cancer, 24(1). https://doi.org/10.1186/s12885-024-12776-8.
- Wang, C. et al. (2022) ‘Presence of Concurrent TP53 Mutations Is Necessary to Predict Poor Outcomes within the SMAD4 Mutated Subgroup of Metastatic Colorectal Cancer,’ Cancers, 14(15), p. 3644. https://doi.org/10.3390/cancers14153644.
- Fang, T. et al. (2021) ‘Prognostic role and clinicopathological features of SMAD4 gene mutation in colorectal cancer: a systematic review and meta-analysis,’ BMC Gastroenterology, 21(1). https://doi.org/10.1186/s12876-021-01864-9.
- Kawaguchi, Y. et al. (2019) ‘Mutation Status of RAS, TP53, and SMAD4 is Superior to Mutation Status of RAS Alone for Predicting Prognosis after Resection of Colorectal Liver Metastases,’ Clinical Cancer Research, 25(19), pp. 5843–5851. https://doi.org/10.1158/1078-0432.ccr-19-0863.